This theme focuses on plant valorization and bacterial interactions to find new ways to fight microbial infection to protect crops and human health. We are also relying on the theme Biomimicry and Biomolecular Diversity to develop new approaches to improve/characterize molecular interactions underpinning our questions.
Knowledge of lipid systems
The increasing demand for plant oils, for nutritional and industrial applications, is the driven theme behind our efforts toward a better characterization of plant lipid metabolism. 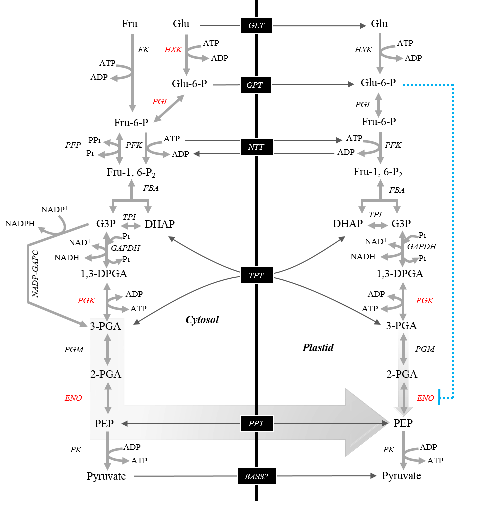 We tackle this challenge from various approaches, from classic biochemistry to “omics” techniques and using different biological systems, from model plants (Arabidopsis thaliana) to staple oilseeds crops. We are also interested on the consequences of climate change upon plant lipid metabolism. Climate changes and global warming modify the distribution of arable land, therefore, deciphering the connections between external inputs, particularly temperature and light, the internal circadian clock and lipid metabolism will contribute to understand the adaptability of oilseeds to different latitudes and to obtain better-fit varieties.
We tackle this challenge from various approaches, from classic biochemistry to “omics” techniques and using different biological systems, from model plants (Arabidopsis thaliana) to staple oilseeds crops. We are also interested on the consequences of climate change upon plant lipid metabolism. Climate changes and global warming modify the distribution of arable land, therefore, deciphering the connections between external inputs, particularly temperature and light, the internal circadian clock and lipid metabolism will contribute to understand the adaptability of oilseeds to different latitudes and to obtain better-fit varieties.
Contact: A. Troncoso-Ponce ![]() B. Thomasset
B. Thomasset ![]() Y. Perrin
Y. Perrin ![]()
Host plasma membrane and bacterial interactions
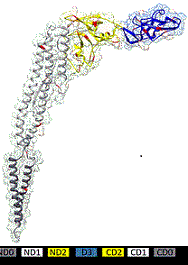 We characterize mechanisms of interactions of bacterial appendages or molecules (glycolipids or peptides) with host plasma membrane (plants and mammals). This allows understanding how bacteria can sense surfaces and will lead to new ways of targeting pathogenic bacteria by identifying mechanisms that may have roles in the initiation of bacterial colonization. We are focusing on molecules having an eliciting (triggering plant defense and/or antimicrobial mechanisms) or antimicrobial effects.
We characterize mechanisms of interactions of bacterial appendages or molecules (glycolipids or peptides) with host plasma membrane (plants and mammals). This allows understanding how bacteria can sense surfaces and will lead to new ways of targeting pathogenic bacteria by identifying mechanisms that may have roles in the initiation of bacterial colonization. We are focusing on molecules having an eliciting (triggering plant defense and/or antimicrobial mechanisms) or antimicrobial effects.
Whole plant valorisation
 Exploitation of whole plant as bioresources to identify and produce economically attractive, renewable and biodegradable, molecules. Possible uses lie in biolubricants, in biofuels and molecules beneficial for human or animal health (antioxidants, anticancer molecules…).
Exploitation of whole plant as bioresources to identify and produce economically attractive, renewable and biodegradable, molecules. Possible uses lie in biolubricants, in biofuels and molecules beneficial for human or animal health (antioxidants, anticancer molecules…).
Contact: Eric Husson ![]() Catherine Sarazin
Catherine Sarazin ![]()
Rational design and optimization
In our researches, in silico approaches are developed to propose original solutions dedicated to binding or inhibition.
Our technological arsenal
Targeted and untargeted lipidomics and fluxomics by mass spectrometry (plants, bacteria, fungus…). Post translational modification characterization. Isotopic profiling. Plant elicitation assays (callose quantification, ROS quantification, stomatal aperture measurement, conductimetry…), transcriptomic analysis, plant physiological effects of elicitors, antifungal assays (rapeseed pathogens) and phytoprotection assays. Arabidopsis mutants (bank mutant screening, crossing), plant recombinant protein production, enzymatic assays. Peptide data mining. Chemical and biophysical characterization of amphiphilic elicitor composition (mass spectrometry, DLS, contact angle). Interaction with biomimetic membrane (solid state NMR, molecular modeling, SPR…). Visualization, quantification and characterization of bacterial adhesion with biotic or abiotic surfaces.
Participants:
B. Thomasset (DR CNRS), E. Ruelland (CR CNRS), Y. Perrin (PR UTC), C. Sarazin (PR UPJV), N. D’Amelio (PR UPJV), R. Figge (Chaire senior UPJV), A. Roscher (MC UPJV), I. Gosselin (MC UPJV), E. Husson (MC UPJV), A. Troncoso-Ponce (Chaire Junior), S. Rippa (IR UTC), S. Acket (IE UTC)
