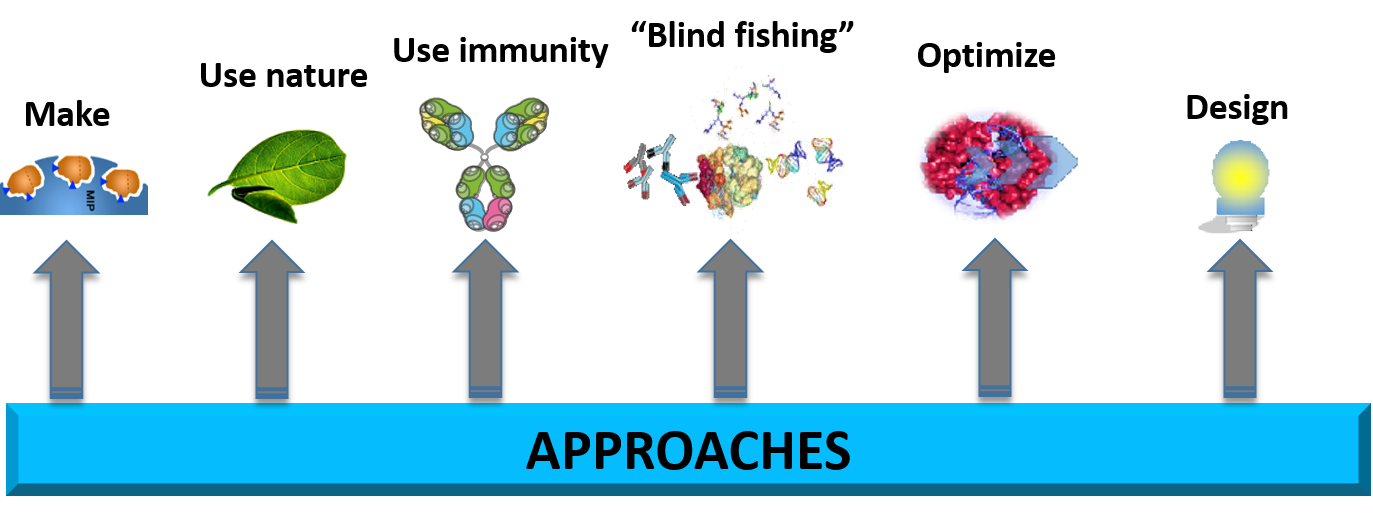
Our research is centered on the premise that solutions to scientific challenges are present in Nature. Our objectives tend toward new molecules inspired by Nature’s patterns to be used in all the fields covered by human activities (chemistry, medicine, cosmetology, agriculture…).
Biomimetic materials
We develop new versatile biomimetic materials with high recognition capabilities for a target molecule based on synthetic polymers called Molecularly Imprinted Polymers (MIP). Molecular imprinting relies on the attractive molecular recognition of the functionalities of a template by the complementary functional groups of monomer(s) anchored in a matrix formed by cross-linked monomers. Extraction of the template leaves in the polymer cavities complementary in shape, size and chemical functionality to the target. The MIP thus obtained binds the analyte with an affinity and specificity comparable to that of natural antibodies and are often referred as antibody mimics or ‘plastic antibodies”.
Biomimetic materials by green chemistry approaches were developed to propose new multi-functional polymers compatible with applications requiring materials with biocompatible, biodegradable or functional properties.
Libraries of biomolecules
 Developments in protein engineering and molecular biology techniques have provided us with the possibility to produce in vitro molecular diversities close to natural immune repertoires. With phage display technology, we attempt to approximate this diversity. Our disposal of 4 antibody libraries based on different murine models, comprising auto-immunity prone mice, allows us to work on libraries that have experienced different maturation processes.
Developments in protein engineering and molecular biology techniques have provided us with the possibility to produce in vitro molecular diversities close to natural immune repertoires. With phage display technology, we attempt to approximate this diversity. Our disposal of 4 antibody libraries based on different murine models, comprising auto-immunity prone mice, allows us to work on libraries that have experienced different maturation processes.
 Libraries of biomolecules such as peptides, aptamers, mutants, are reservoirs of molecular diversity from where any ligands can be isolated using iterative selection processes (SELEX or phage display), without any a priori knowledge of the target. We are able to target proteins, active sites, small compounds, and whole cells.
Libraries of biomolecules such as peptides, aptamers, mutants, are reservoirs of molecular diversity from where any ligands can be isolated using iterative selection processes (SELEX or phage display), without any a priori knowledge of the target. We are able to target proteins, active sites, small compounds, and whole cells.
Contact: Séverine PADIOLLEAU and Bérangère BIHAN-AVALLE ![]()
Natural compounds
Bioactive natural compounds are by definition a source of novel products that may be alternative to synthetic molecules or answers to fundamental or technological issues. We study interactions of natural compounds with extracellular matrixes, cell receptors or plasma membranes as well as their modulatory effects. In order to optimize the bioavailability of these biomolecules for their final valorisation in food or pharmaceutical products, we study their stabilization by encapsulation and their release kinetic under physiological conditions.
The knowledge gained about natural compound activities and nutritional biochemistry is applied to the development of new functional food products through the Food Science platform.
Contact: Mirian KUBO and Claire ROSSI ![]()
Rational design and optimization
Some of our research activities take advantage of in-silico approaches to propose original solutions dedicated to optimal binding or inhibition. They also provide clues for optimization of molecules of interest and interactions (docking) and for the design of epitope mimics.
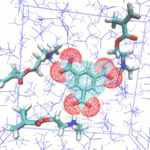
In the field of molecular imprinting, in-silico calculations allow to screen for the best functional monomer in terms of affinity for its template molecule such as to optimize the template-monomer complex formation and therefore the binding capacity of the final MIP. When possible, the in-silico screening studies are cross-validated by experimental titration measurements (NMR, ITC, IR). Such virtual screening approaches are particularly relevant for expensive templates for example.
Peptide Biophysics team
In all living organisms, peptides are used to modulate key biological processes and are part of the innate immune system to fight cancer and infections. In the era of antimicrobial resistance, new antibiotics are urgently needed, while peptides are being rediscovered as drugs. Even though nowadays more and more peptides are in clinical trials, a detailed description of their mechanism of action is required to optimise their activity. Their flexibility and small dimensions prevent the study by powerful structural biology techniques such as X-ray diffraction and cryo-microscopy. However, solid and liquid-state NMR and Molecular Dynamic simulations can reveal their three-dimensional structure and dynamics at atomic level, even when interacting with biological membranes of entire living cells.
Contact: http://gec.u-picardie.fr/gec-amps/
Our technological arsenal
Molecular biology of bacteriophages (cloning, expression, purification); Iterative selection processes (SELEX, phage display); Production of recombinant proteins in prokaryotic and eukaryotic systems; Computational methods; Rational molecularly imprinted polymers; 2-photon stereo lithography; Green chemistry for biocompatible mimetic materials; Screening of molecular interactions experimentally (NMR, ITC) and in silico.
Participants
K. Haupt (PREX UTC), B. Bihan-Avalle (PR UTC), A. Friboulet (DR CNRS), C. Rossi (PR UTC), N. D’Amelio (PR UPJV), F. Ramos-Martín (MC UPJV), I. Gosselin (MC UPJV), R. Figge (Chaire Senior UPJV), C. Sarazin (PR UPJV), S. Padiolleau-Lefèvre (MC UTC), I. Maffucci (MC UTC), S. Octave (IR UTC), A. Falcimaigne-Cordin (MC UTC), C. Gonzato (MC UTC), O. Wattraint (MC UPJV), B. Bouvier (CR CNRS), E. Prost (IE CNRS)